Introducing MolSpin, a remarkable creation born in the heart of our very own research group. MolSpin, which stands for Molecular Spin Dynamics, isn't just your run-of-the-mill software—it's a virtual playground for unraveling the mysteries of spin systems. Think of it as your personal spin dynamics laboratory, where you can dive into the captivating world of spins and explore their behavior in any context you desire.
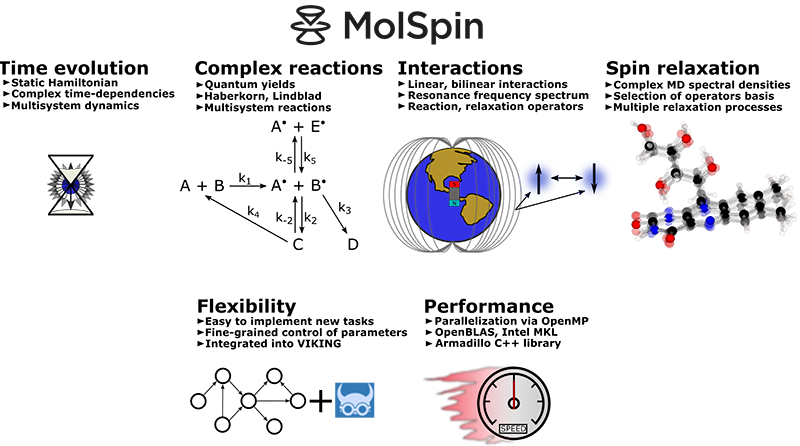
But what sets MolSpin apart is its boundless nature. It is designed to be incredibly versatile, focusing on adaptability. This means that when the standard features don't quite fit the bill, you can easily craft new algorithms to suit your needs, making MolSpin your creative canvas. Forget the hassle of starting with a new script; MolSpin empowers you to extend its capabilities easily, allowing your imagination to roam freely.
And here's the real gem: MolSpin doesn't confine itself to specific types of calculations. While it excels at solving the Liouville-von Neumann equation, it's also a master of versatility. Whether diving into semi-classical simulations or even taking a sneak peek into the mesmerizing realm of quantum computing, MolSpin is your trusty companion, ready to help you explore the depths of spin dynamics in ways you've never imagined. It's not just software; it's a ticket to boundless discovery in the enchanting world of molecular spins.
For more information, please visit https://molspin.eu.
