Integral to many biological functions such as signaling, energy conversion, and cellular interactions, membrane-embedded proteins play a pivotal role in various physiological processes. For instance, rhodopsin in the eyes facilitates vision, and ATP synthase in mitochondria is central to energy production. To understand these membrane-protein complexes, we employ advanced molecular dynamics simulations, offering a comprehensive view of intricate protein movements and functions.
Our approach involves the integration of molecular mechanics with quantum chemical calculations, enabling us to unravel detailed insights into the reaction mechanisms underlying different processes. This is of paramount importance as membrane-embedded proteins are often closely tied to health and disease.
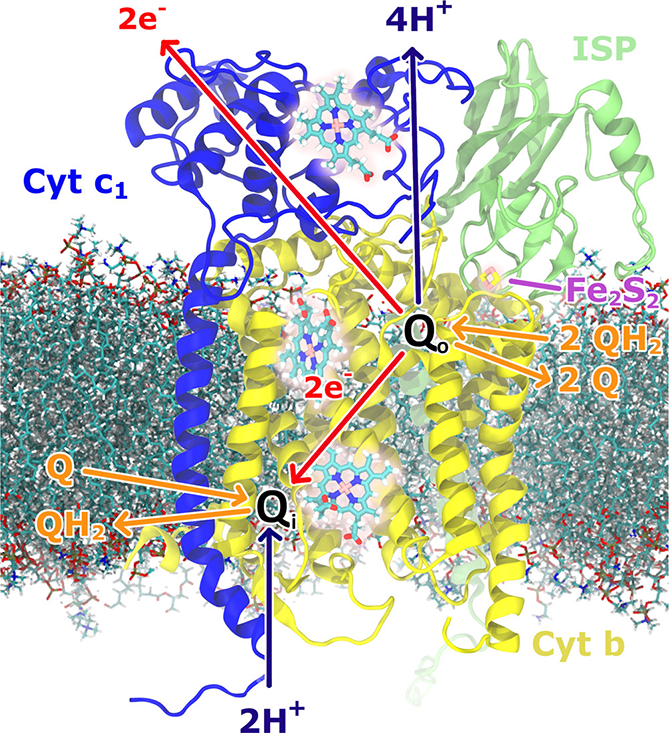
One focal point of our investigations is the cytochrome bc1 complex, a crucial component in cellular respiration and photosynthesis. Through sophisticated simulations, we've uncovered its dynamic role in electron and proton transfers, essential for ATP synthesis. Notable research highlights include examining the complex's interaction with oxygen, potentially leading to harmful superoxide production, and understanding how mutations influence drug resistance in diseases like malaria. These insights are instrumental in developing targeted therapeutics and enhancing our understanding of cellular aging processes.
